VIDEO and FREE TRIAL
Qlucore Omics Explorer video
Qlucore Omics Explorer free trial
Qlucore Omics Explorer is a next-generation bioinformatics software for research in life science, biotech, food and plant industries, as well as academia. The fast and interactive visualization-based data analysis tool with inbuilt powerful statistics delivers immediate results and provides instant exploration and visualization of a wide range of omics data including miRNA.
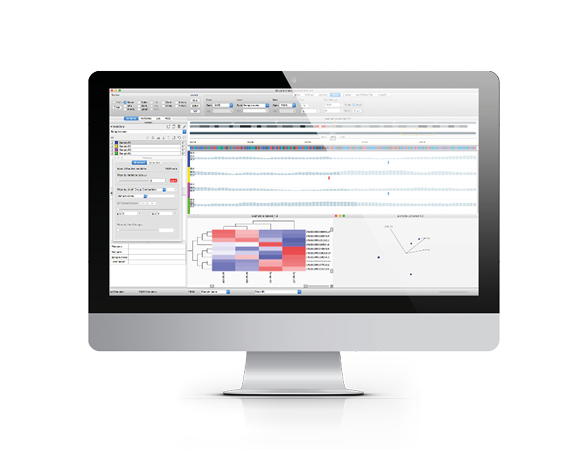
miRNA (also written microRNA or µRNA) are non-coding RNA that are not translated into proteins. Instead they normally control the translation of mRNA. With Qlucore Omics Explorer, a researcher can easily examine and analyze data from miRNA (microRNA) experiments. Data can be generated either by NGS techniques or microarrays.
Answer the four quick questions below and find out if you can use Qlucore on your data.
For more details about supported data formats and data import see Data Import or Contact us with questions.
The study includes working with data from more than 400 arrays. Visualization is used in the effort to understand the human growth process.
Understanding viral signatures in Hepatitis C. Qlucore Omics Explorer is used to identify differences in chemokines between the virally infected groups.