
DNA methylation analysis
Qlucore Omics Explorer is the powerful visualization-based data analysis tool with inbuilt powerful statistics delivers immediate results and provides instant exploration and visualization of big data. The program is well suited as well for analysis of DNA methylation data. The application areas can be cancer or others, the generic approach gives the user freedom to approach a wide spectrum of research questions.
Analyze DNA methylation data
DNA methylation is a type of chemical modification of DNA that can be inherited and subsequently removed without changing the original DNA sequence. As such, it is part of the epigenetic code and is also the best characterized epigenetic mechanism. Research has shown that DNA methylation is manifested in a number of important biological processes and human diseases including cancer. The program is well suited as well for analysis of Illumina DNA methylation.
In Qlucore Omics Explorer a number of different analyses can be done.
Key functionality
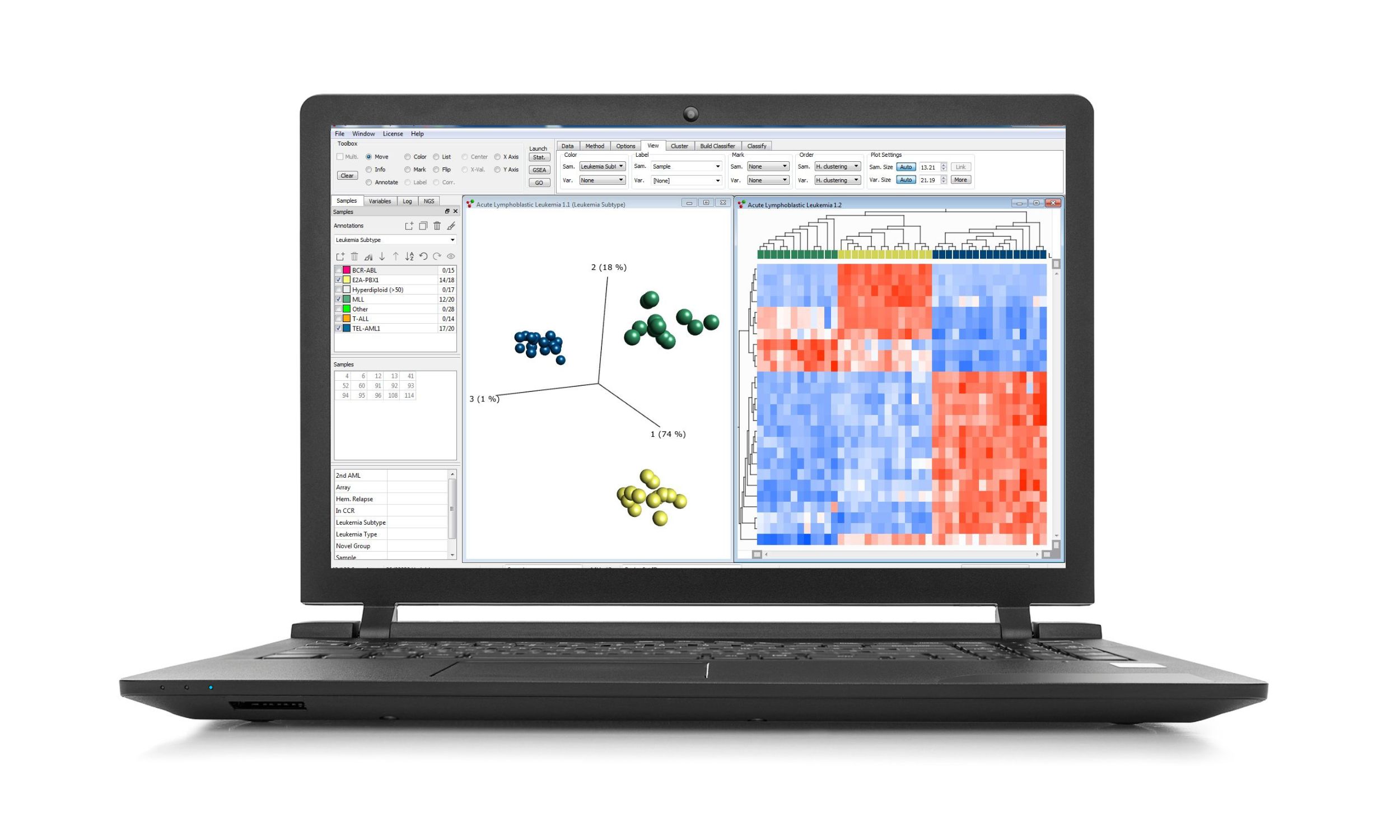
- Check data for outliers by visual inspection using sample Principal Component Analysis (PCA) plots.
- Perform statistical filtering using ANOVA to enhance results. Unwanted dependencies can be removed with one mouse click.
- Generate a list of genes that classifies data based on a selection of statistical tests: f-test, t-tests or regression.
- Present results using for instance heatmaps or PCA plots.
Does it work on my data?
Answer the four quick questions below and find out if you can use Qlucore on your data.
For more details about supported data formats and data import see Data Import or Contact us with questions.
Case Studies
Analysis of public data using Qlucore
Beijing Normal University, ChinaThis case study is an example of how the use of public information from multiple sources was used to propose a new classification for glioma cancer.