VIDEO and FREE TRIAL
Qlucore Omics Explorer video
Qlucore Omics Explorer free trial
Mette K. Andersen (MD, PhD) is a clinical geneticist at the Department of Clinical Genetics at Rigshospitalet which is the largest and most specialized hospital in Copenhagen (Denmark).
She specializes in Clinical Genetics.
“One thing that really stands out is the combination of a gene expression subtype classifier and the detection of gene fusions.”
CURRENT STATE OF CLASSIFICATION IN OUR LAB
In pediatric acute lymphoblastic leukemia (ALL) the acquired genetic profile is important for risk-stratification and choice of therapy. Gene fusions are common, but only the most frequent gene fusions will be detected by standard methods. New gene fusions and other subtypes of diagnostic and prognostic significance may be overlooked by standard genetic methods. One example is that currently only the ABL1 gene is identified in detection of one of the ABL class fusion genes through FISH analysis and not the partner genes. Another concern is that in a number of cases, suspected findings lack a standardized way of confirmation.
In the Rigshospitalet genetic laboratory, 30 cases of ALL are analyzed annually. ALL is one of the most severe diseases in children even if the majority is successfully treated. Selecting the optimal therapy is important. In response to this need the objective has been to implement RNA sequencing in routine diagnostics of pediatric ALL.
SOLUTION
The team at Rigshospitalet has implemented the use of Qlucore Insights for in-house use, in accordance with Article 5(5) in EU IVDR, and included it in the RNA lab workflow. As data input whole RNA sequencing data (WTS) from diagnostic ALL samples are used.
The Qlucore Insights software, which is intended for research use only, has been validated for in-house use by the team at Rigshospitalet, by comparing the results of RNA sequencing with standard methods such as G-band karyotyping, FISH, SNP array and RT-PCR. Forty-six B-ALL samples and 8 T-ALL samples from the biobank have been tested. Based on the validation results, RNA sequencing data analysis using Qlucore Insights is now incorporated as a routine diagnostic tool.
Of the 54 samples in the validation cohort, all known findings were confirmed except in one case. As examples of new findings, the classifier in Qlucore Insights identified a DUX4 rearrangement in a B-ALL sample and a suspected MLLT10::PICALM gene fusion was confirmed in a T-ALL sample. Thus, correct genetic diagnostics was provided using the software Qlucore Insights for Research Use Only.
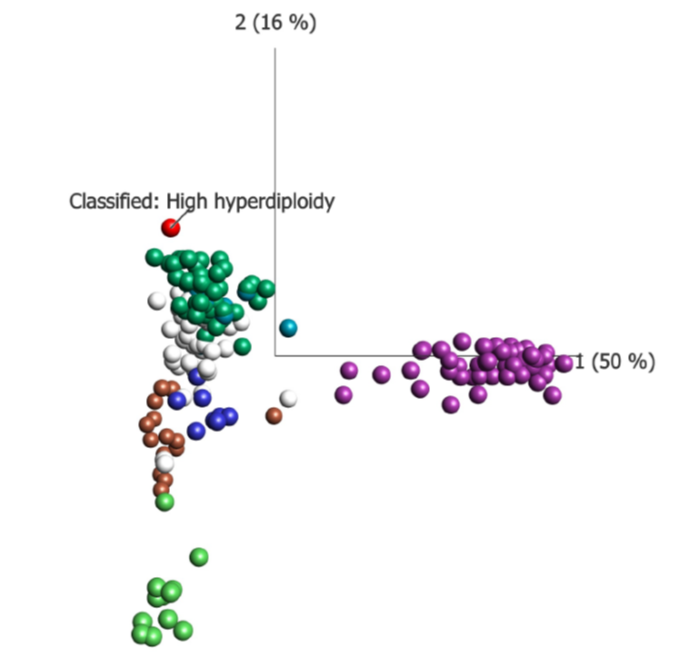
Figure: A PCA plot illustrating how a patient classified as subtype High hyperdiploidy relates to the reference data set. As can be seen the red dot is close to the green reference samples in the green group – High hyperdiploidy.
“Other labs should use Qlucore Insights to improve pediatric genetic ALL diagnostics.”
A great advantage of the Qlucore Insights software is that it not only reports all fusion genes, but based on gene expression profiles provides information about common, rare and new subtypes of B-ALL. The included subtypes are high hyperdiploidy, BCR::ABL or BCR::ABL like, DUX4 rearranged, ETV6::RUNX1 or ETV6::RUNX1 like, KMT2A rearranged, and TCF3::PBX1. See figure below.
As mentioned, the validation has been successful and the introduction in routine in-house diagnostics at Rigshospitalet has worked very well.
The information in the report includes both quality parameters, visualizations and probabilities for subtype likelihood. This is very useful for communication. See example below.

Qlucore Insights can be used to improve genetic analysis in pediatric ALL. It has the potential to complement other methods, particularly to confirm findings and when other methods fail. Another advantage is that one advanced analysis based on whole transcriptomics sequencing(WTS) may replace some of a whole set of the traditional genetic tests otherwise required.